
DOUGHERTY LAB
From Astrocytes to Autism
Our laboratory is interested in the genetics and genomics of behavior in health and disease. We utilize a variety of techniques, including human molecular genetics, informatics, mouse behavior, in vitro and in vivo neuroscience, and neuroanatomy. We are continuously developing novel methods for transgenesis, gene manipulation, and transcriptional profiling of the brain. These tools help us to develop mouse models for discovery and modeling of genetic variations based on human patient populations, in order to understand the cellular and molecular underpinnings of behavior. We are particularly focused on neurodevelopmental conditions, including the autism spectrum.
We study intellectual and developmental syndromes to better understand how gene expression differences can change the trajectory of neural development. We believe that a better understanding of the basic science behind syndromic conditions has the potential to give patients and families more agency and options in addressing their unique challenges. We also are very interested to understand and be informed by the needs of the families themselves, and welcome engagement with the community of the syndromes we work on. Our specific applications of these methods currently span six different, inter-related areas of research:
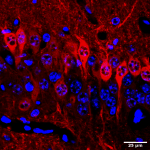

Transcriptional and translational regulation, and its dysregulation in neuropsychiatric and neurodevelopmental conditions:
- Impacts of genetic variation. Genetic variants, both those common in the population and those which are de novo—which are variations not seen in the parents but found in an offspring, due to e.g. spontaneous mutations in germ cells—have been associated with neuropsychiatric diseases by way of genome-wide association studies and genomic analysis of parent-child trios, respectively. In both cases, a vast number of the variations associated with or unique to disease fall in non-protein coding DNA, making it difficult to discern whether and how a variation begets disease. Using Massively Parallel Reporter Assays (MPRAs), we can systematically assess the effects of large sets of variants falling in a) untranslated mRNA elements, as well as b) noncoding variants falling in putative regulatory regions. We are also using this technology to explore putative transcriptional regulators of CNS cell types, sex differences in enhancer element usage, potential antisense oligo (ASO) therapy for use in treating intellectual and developmental disability,and the effects of non-coding variants on translation and mRNA stability. We also have been performing ongoing computational genomic analysis in collaboration with the Simons Simplex Consortium, examining mutational burdens and their potential functions in family cohorts with a child suffering from ASD.
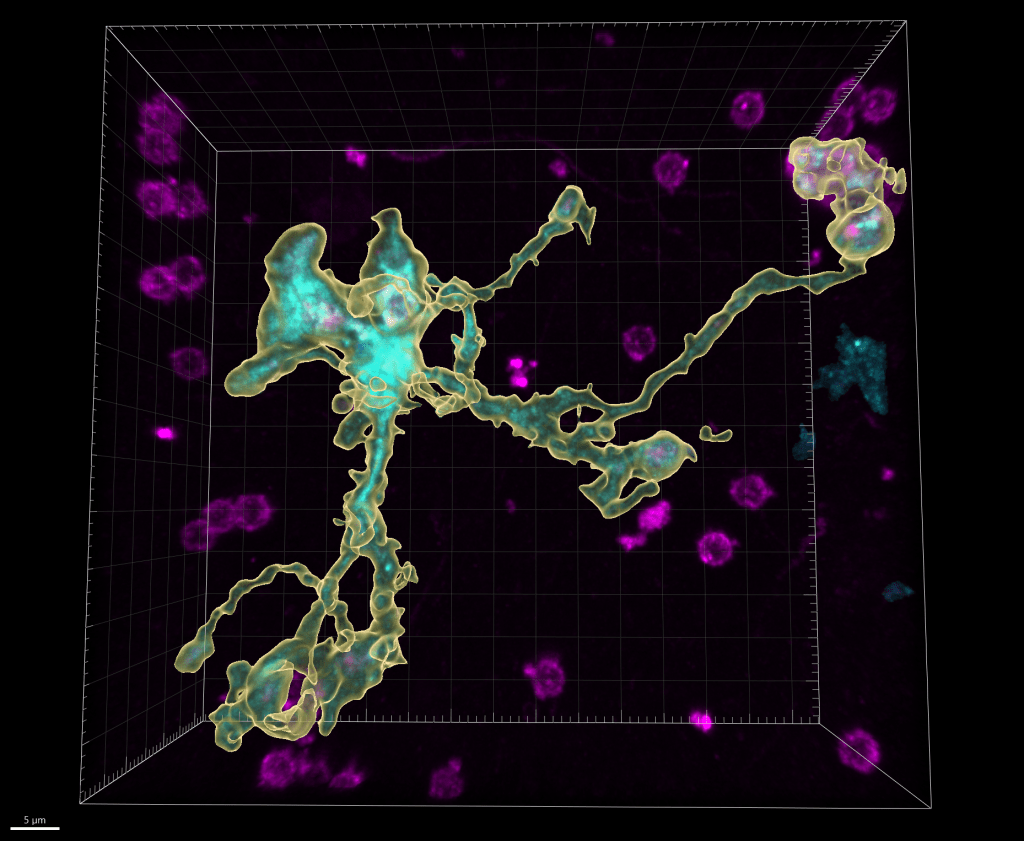
- Local and conditional translation of mRNA. Neurons have been long known to localize certain mRNAs to their axons, dendrites, and synapses for translation at the site of use. Our lab was the first to definitively show and characterize this process in astrocytes, another abundant brain cell type, and microglia, an immune cell of the brain. Using similar techniques, including Translating Ribosome Affinity Purification (TRAP) and Ribosome Footprinting (RF), we are continuing to characterize the mechanisms regulating RNA localization and translation in glial cell types and specific neuronal populations. Moreover, these methods enable us to look at e.g. effects of neuronal stimulation on mRNA occupancy, providing potential insights into mechanisms of conditional translation. Finally, using technologies such as CLIP-seq (an RNA analogue of ChIP seq), one can identify RNA binding proteins and their genes, providing another means of insight into regulation of RNA location and translation.
- Longitudinal transcriptional signatures that underlie individual variance and neuronal disorder phenotypes. Currently available transcriptional tools are limited in that they only provide a one-dimensional snapshot in time – the molecular signature of the brain at the moment of harvest. We sought to overcome this limitation by developing in vivo Calling Cards, a tool that permanently records transient protein-DNA interactions, that builds on a technology pioneered by our collaborators, the Mitra Lab. We further adapted the tool to be compatible with standard single cell technologies, allowing us to assay gene expression and transcription factor binding with single cell resolution. Calling Cards has opened the door to multiple avenues of investigation: characterizing potentially aberrant molecular signatures in models of neurodevelopmental conditions, adapting our technology to only record during specified time windows, and identifying how early-life transcriptional changes can underpin later-life susceptibility or resilience.

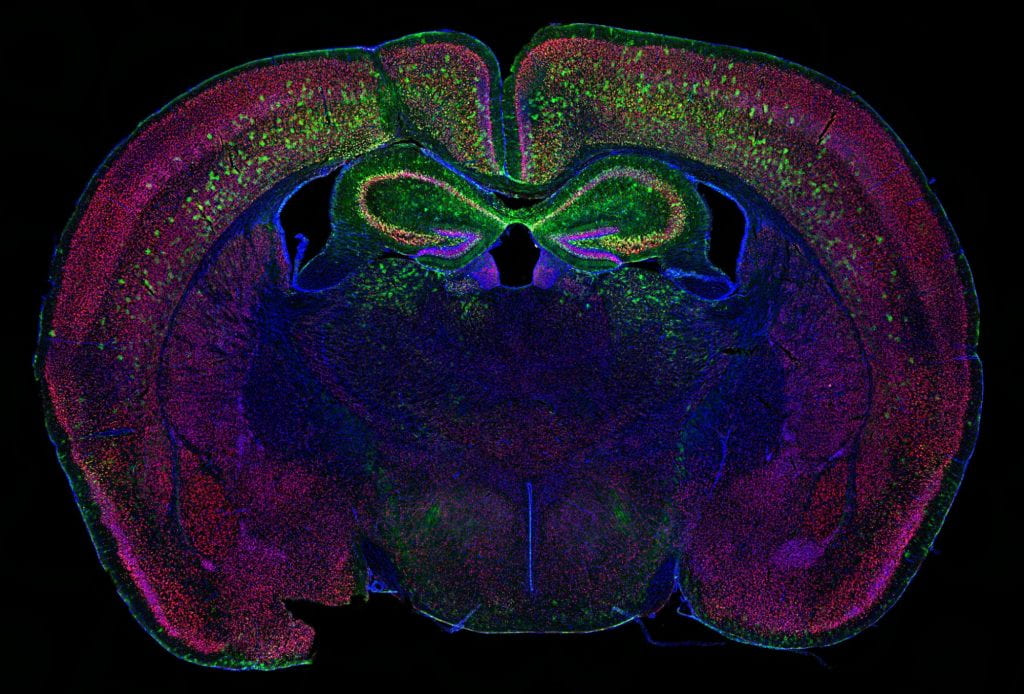
Genetic and behavioral mouse models of neurodevelopmental conditions and risk factors :
- Genetic factors. While one type of genetic variation, discussed above, does not directly impact the protein-coding genome, some variations are capable of compromising one or multiple proteins, especially in the cases of chromosomal mutations and transcriptional factor mutations. In these cases, a gene or a set of genes become implicitly responsible for the observed phenotypes, which makes the disease amenable to modeling by use of gene knockout mice. With the expertise from characterizing mouse models of William Syndrome (a disease that features hypersociability, in interesting contrast to ASDs), our lab continues this journey by focusing on a neurodevelopmental disorder (NDD) related gene, Myelin Transcription Factor 1 Like (MYT1L). Utilizing the MYT1L haploinsufficiency mouse model mimicking a patient mutation, we are seeking to understand how MYT1L mutations lead to various human patient phenotypes from molecular, to cellular, and to circuitry levels. This project leverages a wide range of techniques we established in the lab, including RNA-seq, ATAC-seq, CUT&RUN, MPRA, Calling Card, electrophysiology, and behavioral paradigms. Furthermore, we generated MYT1L conditional knockout (cKO) and conditional rescue mouse lines to investigate cell-type/development specific functions of MYT1L and explore therapeutic opportunities for MYT1L Syndrome.
- Sex differences. Almost all neuropsychiatric diseases appear to carry uneven incidence among males and females—for example, ASC, ADHD, and schizophrenia are diagnosed more in men, while depression, anxiety, and PTSD tend to affect more women. Animal research, as both a behavioral model and a living experimental model of the nervous system, has until recently—and often still does—only performed experiments in male animals. This has left huge gaps in knowledge about whether and how sex affects behavior, neuroanatomy, and gene expression. We have characterized and are continuing to explore the genetic and hormonal mechanisms of sex differences, specifically in depression and autism.
- Environmental factors. Many neurotransmitter and neuromodulator systems targeted by drugs play vital roles in neurodevelopment, and disruption of these systems during critical periods of brain development can have lasting impacts on behavioral circuits. To understand the potential impact of changes to the serotonergic system during neurodevelopment on later behavioral function, , we investigated the role of in utero exposure to selective serotonin reuptake inhibitors (SSRI, a class of antidepressant) on social and sensory behaviors, which are often impacted in intellectual and developmental disabilities. We found that this early targeting of the serotonin system resulted in reduced early social communication, altered social hierarchy behaviors, and increased sensitivity to tactile stimulation. We then took the strategies developed to understand SSRI exposure and applied them to develop a model of Neonatal Opioid Withdrawal Syndrome to understand the impact of clinically-relevant continued postnatal opioid maintenance and mitigation strategies on long-term behavioral function. This collaborative effort with the Al-Hasani Lab revealed that ontogenetic exposure to oxycodone disrupts weight trajectories, early communicative behavior and early somatosensory reflexes in offspring, as well as sex-specific changes in sensory and reward processing.
We are endlessly grateful to the institutions and foundations whose funding has made and continues to make our work possible:













