Ushiki Lab | Google Scholar Profile
Bio
Aki earned her PhD from the University of Tsukuba, Japan, under Dr. Keiji Tanimoto, where she studied Igf2/H19 genomic imprinting and transcriptional regulation of the Renin gene using hypertensive and pregnancy-induced hypertensive mouse models. She also gained hands-on experience in various transgenic techniques, including pronuclear injection, gene targeting in ES cells, and CRISPR/Cas9 genome editing.
Subsequently, Aki pursued postdoctoral research in Dr. Nadav Ahituv’s lab at UCSF. Her research primarily focuses on skeletal development and diseases, with a special emphasis on enhancers. She has significantly advanced the understanding of genetic mechanisms underlying congenital limb deformities, adolescent idiopathic scoliosis (AIS), and bat wing development, using genomics and mouse genetics (see publications).
Additionally, she is developing high-throughput enhancer assay technology in mice. Her current research is supported by NHGRI K99 funding.
Awards and Honors
- 2023: NHGRI Pathway to Independence Awards (K99/R00)
- 2020: Reviewers’ Choice (Top 10% of all poster abstracts), American Society of Human Genetics (ASHG) 2020 Virtual Meeting
- 2020: Semi-Finalist, ASHG/Charles J. Epstein Trainee Awards, American Society of Human Genetics (ASHG) 2020 Virtual Meeting
- 2019: JSPS Overseas Research Fellowship
- 2018: Uehara Memorial Foundation Postdoctoral Fellowship
- 2017: Dean’s Award for Excellence, Graduate School of Life and Environmental Sciences, University of Tsukuba, Japan
- 2016: JSPS Research Fellowship for Young Scientists (DC2)
- 2012: Valedictorian Award, College of Biological Sciences, University of Tsukuba, Japan
Research
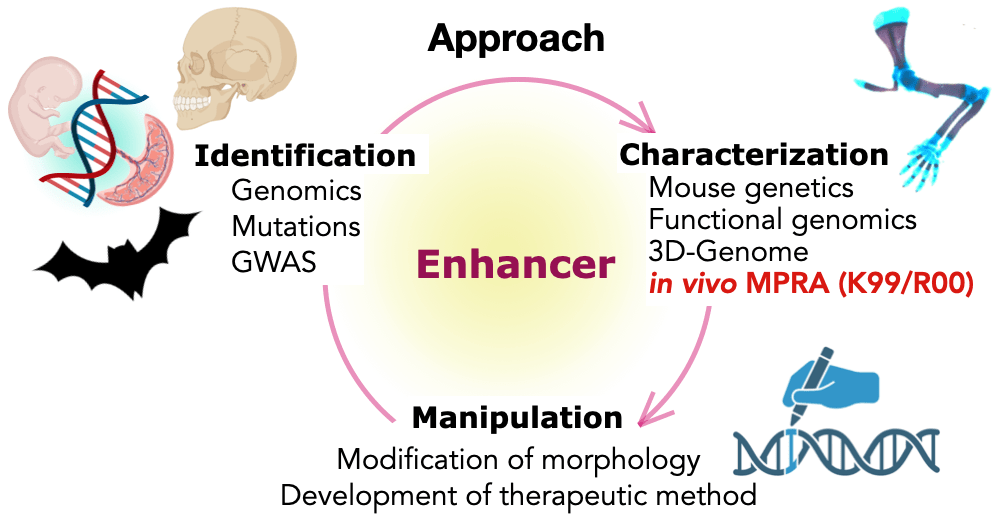
Our lab focuses on understanding how gene-regulatory elements control gene expression in mammals, particularly in relation to the genetic basis of development, evolution, and disease. To study enhancer function, our approach includes three key components (see figure below).
First, we identify putative enhancers from comprehensive genomic datasets and GWAS data. Next, the in vivo functions of these enhancer candidates are characterized using a combination of mouse genetics and functional genomics techniques. Finally, by integrating this functional information, we use enhancers as tools to manipulate phenotypes, such as altering morphology or developing new therapeutic approaches.