From deciphering underlying genetic factors of diseases to developing cutting-edge genome technology, our scientists are making impactful discoveries everyday.
Strength in Genetics and Genomics Research
The Department of Genetics has traditional strengths in computational biology and genome science, as well as model organism, evolutionary and human genetics. Recent specialties include neurological disorders, cellular bioenergetics, epigenomics, personalized medicine and genome technology development.
We have established leadership in the following flagship NIH genomic medicine themed projects:
- The Human Pangenome Project (NHGRI)
- The Impact of Genetic Variation on Function (NHGRI)
- The Long Life Family Study (NIA)
- Somatic Mosaicism across Human Tissues (NIH Common Fund)
- Multi-Omics for Health and Disease (NHGRI, NCI, NIEHS)
- The BRAIN (The Brain Research through Advancing Innovative Neurotechnologies) Initiative Cell Atlas Network (NIMH)
Within the close-knit research community of Washington University School of Medicine, our scientists are supported by a strong foundation. School of Medicine Facts & Figures
| #2 NIH Funding (2023) | $838.3 Million Research Funding 2022 | 19 Nobel Laureates |
Latest News
The Turner Lab Develops HAT to Call De Novo Variants for Short-read and Long-read Sequencing Data
The Turner Lab has recently released Hare And Tortoise (HAT), an automated de novo variant (DNV) detection workflow for highly accurate short-read and long-read sequencing data. The method was published in Bioinformatics in January 2024.
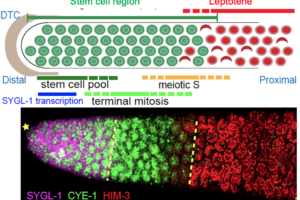
The Schedl Lab Receives R35 Grant
The Schedl Lab led by Dr. Tim Schedl recently received NIGMS R35 grant. The grant provides funding for studying “control of germline stem cells and the switch to meiotic development in C. elegans” for 5 years.
New computational tools developed to identify TE-derived antigens in cancer using long-read CAGE sequencing data
In the new study published in Genome Research, postdoctoral fellow Ju Heon Maeng has developed a suite of computational tools to significantly improve immunopeptidome detection from transposable element expression, utilizing long-read data.