As part of NIH’s BRAIN Initiative, Nature recently published 10 papers mapping the first mammalian brain. Genetics Assistant Professor Dr. Yang (Eric) Li, a contributor in this project, co-first-authored one of the Nature papers titled: Single-cell analysis of chromatin accessibility in the adult mouse brain. Li has also contributed to two other papers in this package, titled: Single-cell DNA methylome and 3D multi-omic atlas of the adult mouse brain, and Conserved and divergent gene regulatory programs of the mammalian neocortex.

Li’s research focused on the examination of chromatin accessibility in 2.3 million individual brain cells from 117 anatomical dissections. The atlas includes approximately 1 million cCREs and their chromatin accessibility across 1,482 distinct brain cell populations, adding more than 446,000 new cCREs to the most recent annotation in the mouse genome. The researchers deduce the gene regulatory networks for over 260 subclasses of mouse brain cells, employing deep learning models to forecast the behaviors of gene regulatory elements in distinct brain cell types solely based on the DNA sequence.
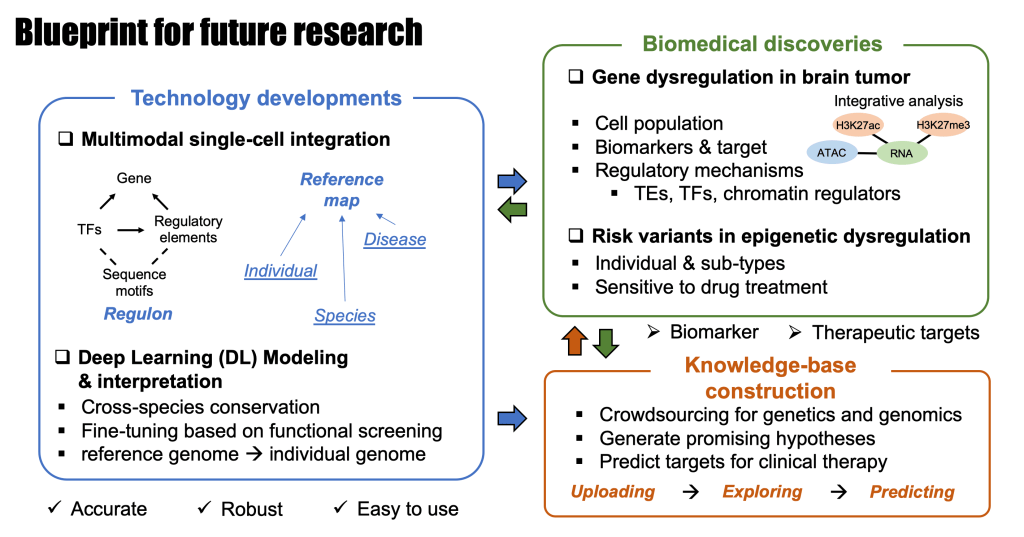
Li started his work on gene regulation in mouse and human brain when he joined Dr. Bing Ren’s lab in the Ludwig Institute for Cancer Research as a postdoctoral researcher in 2018. He is now an Assistant Professor of Genetics and Neurosurgery in Washington University School of Medicine in St. Louis. His lab focuses on developing innovative computational tools/methods and utilizing single-cell (epi)genomic techniques to gain a comprehensive understanding of gene regulation in mammalian models and human diseases, particularly in brain tumors and neuropsychiatric disorders.