Regulatory DNA elements are made of transcription factor binding sites, however how different combinations of binding sites control regulatory DNA to be activating, repressive or having no function at all remains unknown to scientists. In this paper, recently published in PLOS Computational Biology, Dr. Michael White, Associate Professor of Genetics and colleagues used a new AI learning package to model data generated with synthetic regulatory DNA elements to further the understanding of regulatory DNA.
Regulatory DNA is like an alphabet that we haven’t deciphered. “It is like an unknown language where we’ve identified different letters (known binding sites), however we don’t know which arrangements of the letters spell out meaningful words, and which arrangements of letters are nonsense.” said Dr. White. Using deep learning AI models to decipher regulatory elements is advantageous because machine learning can find patterns in data that could be missed by simple statistical analysis. Kai Loell, a recently-graduated PhD student, used the state-of-the-art AI package MAVE-NN developed by colleagues at the Cold Spring Harbor Laboratory to model data generated using regulatory DNA made up of only two types of binding sites in different combinations. The model uncovered a very simple set of “spelling rules”.
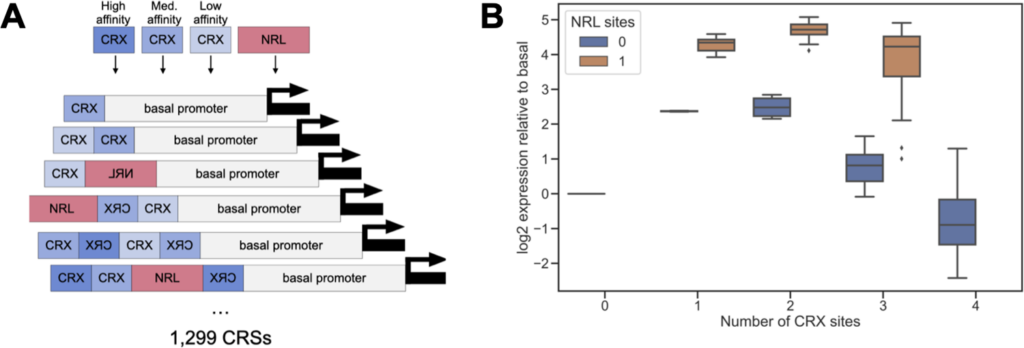
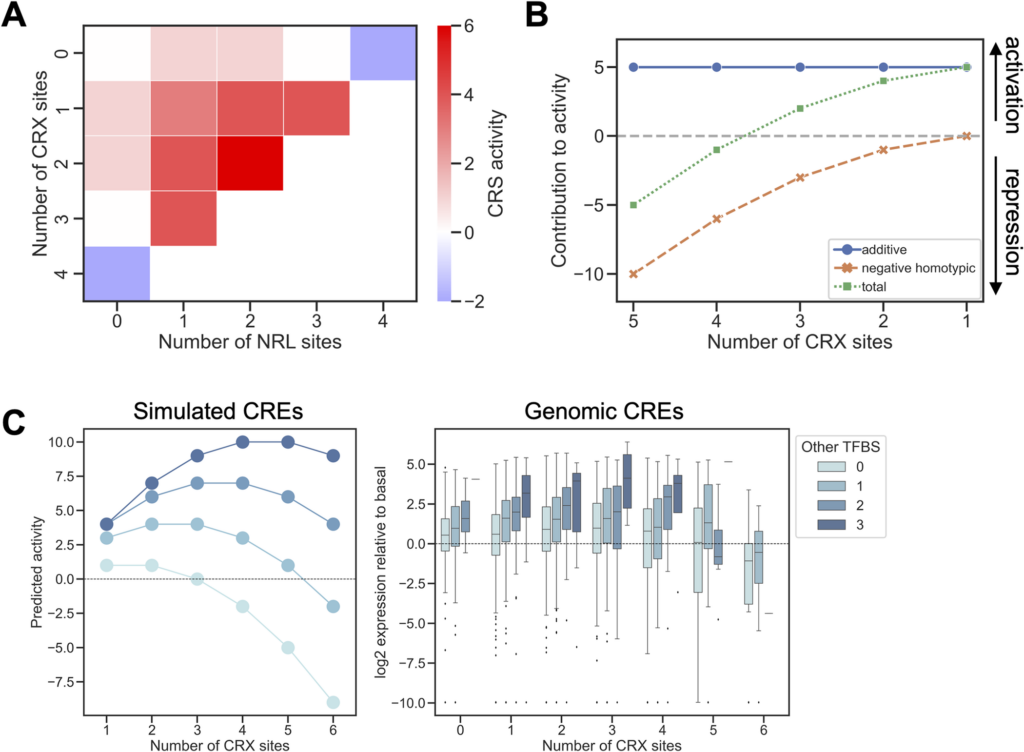
The scientists of this research found that each binding site by itself increases gene expression but when putting two binding sites of the same kind next to each other, it has an inhibitory effect. If you put two different binding sites together, they have a synergistic effect, and it increases gene expression.
After learning the rules from the artificial system created in developing mouse retina cells, the research team then tested these rules on a new data set with 5 different kinds of binding sites as well as a set of natural genomic regulatory DNA sequences. The scientists found that these three simple rules can explain a lot of what’s happening when there are different combinations of binding sites that affect gene activity.
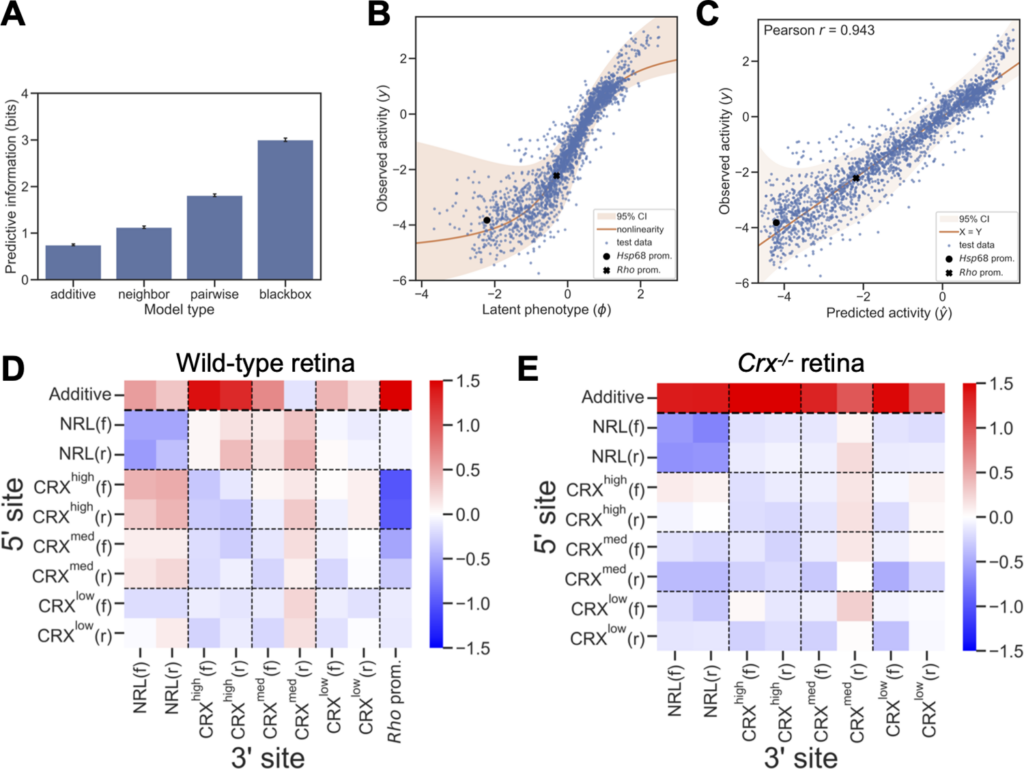
“One significance of this study is that we used a machine learning approach that is interpretable. We can take the machine learning predictions and then boil that down to these three simple rules that the human brain can comprehend.”
Dr. Michael White
Deciphering regulatory DNA in the genome is the main theme of the White and Cohen laboratories. Since a big fraction of the genome is regulatory DNA, disease causing genetic variants and mutations that occur in regulatory DNA are difficult to understand. The ultimate goal of the labs’ work is to learn how to read regulatory DNA and be able to predict what would happen when a genetic variant changes binding site for a transcription factor. It is an important part of understanding how gene regulation affects human health as well.
Featured in this Article
Dr. Michael White, Associate Professor of Genetics

Dr. Michael White obtained his PhD in biochemistry at the University of Rochester, where he developed methods for functional and structural genomics of yeast membrane proteins. He did his postdoctoral work on gene regulation in Barak Cohen’s lab at Washington University in St. Louis. His current work focuses on studying gene regulation using synthetic and natural genomic elements and using deep learning models to model regulatory DNA.
Cohen and White Labs
The Cohen and White labs investigate regulatory DNA by utilizing a multidisciplinary approach, blending genetics, genomics, biophysics, and computational sciences. The lab also aims to develop quantitative models for identifying regulatory sequences, predicting mutation impacts, and enhancing our understanding of their roles in development and disease.